Innovate age-old breeding paradigms
The size control of multicellular organisms is an old biological question that has always fascinated scientists. At present, the question is still far from being solved because multiple molecular pathways are necessary for the formation of a mature organ and their interplay is highly complex. Our long-term goal is therefore to decipher the molecular networks and gene combinations that contribute to agronomic traits.
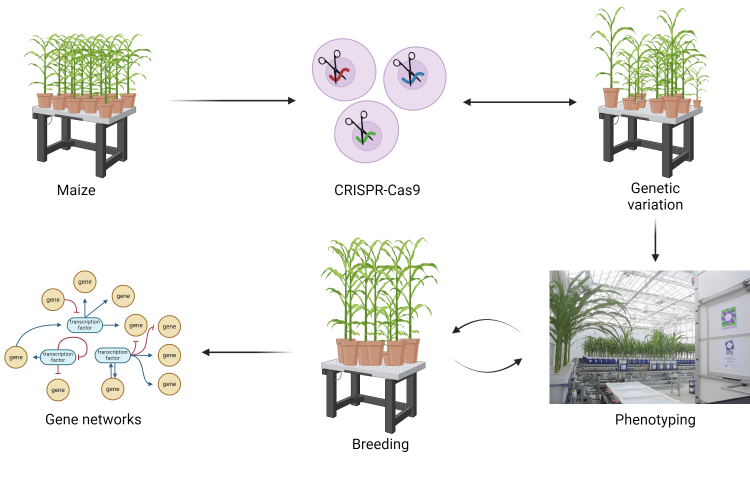
To narrow down the gene space for a given trait and to rationalize which genes to combine, we developed a flexible pipeline called BREEDIT (an acronym of breeding and gene editing) that combines multiplex gene editing with a crossing scheme that integrates higher order edits per generation. Using a Cas9-expressing maize line called EDITOR, super-transformed with multiple guide RNAs (gRNAs) bearing constructs called SCRIPTs, multiple candidate genes can be simultaneously edited and higher order edits can be combined by crossing plants carrying the different SCRIPTs. While a lot of research is done on individual genes that influence leaf size, we are investigating the connections between the various molecular players of the different processes that drive leaf growth to build an integrated growth regulatory network. To increase the effectiveness in finding complex gene combinations that influence maize growth, our team is combining "classical" breeding with multiplex genome editing of growth-related genes identified by investigating mode of action of growth-promoting genes through -omics data. This strategy allows to target entire gene families or multiple components of regulatory pathways, bypassing redundancy and enabling the detection of novel additive or synergistic gene combinations with the aim to improve agronomically relevant traits. Besides a limited number of initial transformations, all other steps solely rely on genetics and genotyping rendering BREEDIT a cost-effective method to scan the gene space for novel combinations of alleles that improve plant traits.